layout: true background-image: url(img/course-logo.png), url(img/logo_SBR.png), url(img//NTU-Logo-full-colour.png) background-position: right top 30px, right 50px bottom 50px,left 50px bottom 50px, top 350px left 500px background-size: 45%, 25%, 20% # Fundamental of Data Science for EESS --- <br> <br> <br> <br> ## R session 07 - Time series .font120[**Daniel Vaulot**] 2020-04-13 --- layout: false class: middle, inverse # Outline .font150[ * Time series in earth sciences * Phytoplankton time series * Manipulating dates: lubridate package * Time series objects: tsibble and feasts packages * Other time series analysis strategies ] --- exclude: true --- background-image: url(img/time_series-book-forecasting.png) background-position: left 300px top 100px background-size: 25% layout: false # Installation and Resources .pull-left[ ## Packages ### New * lubridate * tsibble * feasts * tsibbledata * slider * fable * zoo * WaveletComp ### Previously used * ggplot2 * stringr * dplyr ] .pull-right[ ## Reading list ### Book * Forecasting in practice Chapters 1-4: https://otexts.com/fpp3/ ### Paper * Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329. ] --- layout: true # Time series in Earth Sciences --- background-image: url(img/time_series-paleo_temp.png) background-position: center middle background-size: 80% ## Paleo-temperatures --- background-image: url(img/time_series-paleo_temp_1000.png) background-position: center middle background-size: 70% ## Paleo-temperatures --- background-image: url(img/time_series-Hawaii-CO2.png) background-position: center middle background-size: 50% ## CO2 in the atmosphere --- background-image: url(img/time_series-HOT-CO2.png) background-position: center middle background-size: 50% ## CO2 in the atmosphere in the ocean --- background-image: url(img/time_series-sismo.png) background-position: center middle background-size: 50% ## Seismic records --- background-image: url(img/time_series-lynx-02.png) background-position: center middle background-size: 50% ## Ecology --- layout: true # Marine phytoplankton bloom (_Synechoccus_) --- background-image: url(img/2016-Hunter-title.png), url(img/2016-Hunter-time-series.png) background-position: right 20px top 20px, left 100px top 100px background-size: 30%, 80% --- exclude: true background-image: url(img/flow-cytobot-03.jpg) background-position: bottom 50px center background-size: 45% ## Flow Cytobot * Diatoms --- ## _Synechococcus_ * Discovered in 1979 - _Epifluorescence microscopy_ .pull-left[ <img src="img/Synechococcus_BIOSOPE_Microscopy_3665.jpg" width="70%" style="display: block; margin: auto;" /> ] .pull-right[ <img src="img/1979-Waterbury-fig1.png" width="70%" style="display: block; margin: auto;" /> ] .footnote[_Waterbury, J.B., Watson, S.W., Guillard, R.R.L. & Brand, L.E. 1979. Nature. 277:293–4._] --- background-image: url(img/flow-cytobot-01.jpg), url(img/flow-cytobot-02.png) background-position: bottom 200px left 50px, bottom 50px right 100px background-size: 30%, 60% ## Flow Cytobot * Imaging and flow cytometry --- background-image: url(img/binary_fission.gif) background-position: center middle background-size: 45% ## Cell multiplication * Binary fission * Typically once every day --- background-image: url(img/Synechococcus_phage.png), url(img/Synechococcus_predation_dino.png) background-position: bottom 50px left 150px, bottom 100px right 100px background-size: 20%, 40% ## Cell disappearance * Virus * Predation * Cell death (UV, nutrient deprivation) --- exclude: true ## Growth rate vs Loss rate <br> <br> .pull-left[ `\(\dfrac{\mathrm{d}N}{\mathrm{d}t}=\mu_{net}*N\)` `\(N = N_{0}\exp^{\mu_{net}*t}\)` `\(\mu_{net} = \mu_{growth} - \mu_{loss}\)` * Growth rate = division * Loss rate = cell death, predation, viruses ] .pull-right[ 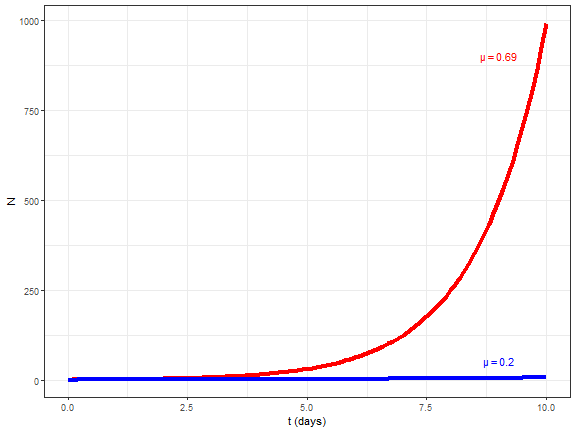<!-- --> ] --- exclude: true ## Growth rate vs Loss rate <br> <br> .pull-left[ `\(\dfrac{\mathrm{d}N}{\mathrm{d}t}=\mu_{net}*N\)` `\(N = N_{0}\exp^{\mu_{net}*t}\)` `\(\mu_{net} = \mu_{growth} - \mu_{loss}\)` * Growth rate = division * Loss rate = cell death, predation, viruses ] .pull-right[ 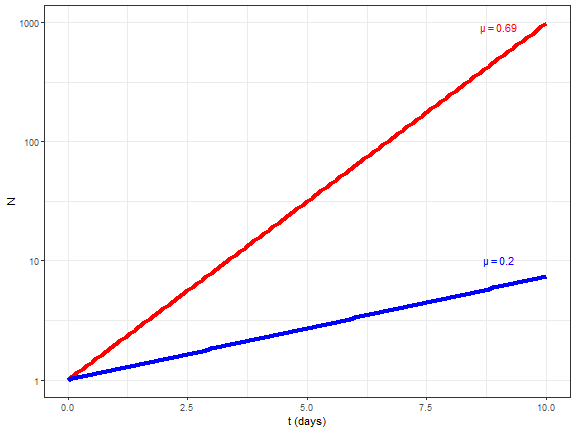<!-- --> ] --- background-image: url(img/2016-Hunter-Syn-01.png) background-position: bottom 50px center background-size: 55% ## _Synechococcus_ abundance .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- exclude: true background-image: url(img/2016-Hunter-Syn-02.png) background-position: bottom 50px center background-size: 55% ## _Synechococcus_ abundance .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- background-image: url(img/2016-Hunter-Temp.png) background-position: bottom 50px center background-size: 55% ## Temperature .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- exclude: true background-image: url(img/2016-Hunter-Temp-02.png) background-position: bottom 150px center background-size: 55% ## Temperature anomaly .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- exclude: true background-image: url(img/2016-Hunter-timing.png) background-position: bottom 50px center background-size: 45% ## _Synechococcus_ vs. Temperature .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- exclude: true background-image: url(img/2016-Hunter-loss_division_spring.png) background-position: bottom 80px center background-size: 65% ## Loss _vs._ Division rate .footnote[Hunter-Cevera et al. 2016. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 354:326–329.] --- layout: true # Manipulating dates: lubridate package --- background-image: url(img/lubridate.png) background-position: center middle background-size: 60% .footnote[https://lubridate.tidyverse.org/] --- ## Parse date Dates are often read as character from files. In order to be recognized in R as dates, they must be parsed .pull-left[ ```r library(lubridate) (bday <- ymd(19530822)) ``` ``` [1] "1953-08-22" ``` ```r (bday <- dmy("22-08-1953")) ``` ``` [1] "1953-08-22" ``` ```r (bday <- dmy("22 aug 1953")) ``` ``` [1] "1953-08-22" ``` ] --- ## Parse date .pull-left[ ```r bday <- dmy(22-08-1953) ``` ``` Warning: All formats failed to parse. No formats found. ``` .warning[do not forget the quotes] ## Parse date and time ```r (bday_time <- dmy_hm("22-08-1953 16:00")) ``` ``` [1] "1953-08-22 16:00:00 UTC" ``` ] --- ## Extract information from dates .pull-left[ ```r bday <- dmy("22-08-1953") year(bday) ``` ``` [1] 1953 ``` ```r month(bday) ``` ``` [1] 8 ``` ```r day(bday) ``` ``` [1] 22 ``` ```r wday(bday, label=TRUE, abbr = FALSE, week_start = 1) ``` ``` [1] samedi 7 Levels: lundi < mardi < mercredi < jeudi < vendredi < ... < dimanche ``` ] --- ## Rounding up dates .pull-left[ ```r # Rounding to start of the month floor_date(bday, unit="month") ``` ``` [1] "1953-08-01" ``` ] --- ## Arithmetic .pull-left[ ```r # Adding one month bday + months(1) ``` ``` [1] "1953-09-22" ``` ```r # Adding years bday + years(66) ``` ``` [1] "2019-08-22" ``` ] --- ## Intervals .pull-left[ ```r # Creating intervals (binterval <- lubridate::interval(bday, Sys.Date())) ``` ``` [1] 1953-08-22 UTC--2020-04-09 UTC ``` ```r # Number of seconds in interval int_length(binterval) ``` ``` [1] 2102716800 ``` ```r # Number of days seconds_to_period(int_length(binterval)) ``` ``` [1] "24337d 0H 0M 0S" ``` ] --- layout: true # Manipulating time series: tidyverts packages --- background-image: url(img/time_series-tidyverts.png) background-position: center middle background-size: 60% .footnote[https://tidyverts.org/] --- ## Loading necessary packages .left-code[ ```r library(tidyverse) library(lubridate) # Time series manipulation library(tsibble) # Time series graphics and statistics library(feasts) # Moving computations library(slider) # Forecasting functions library(fable) # Interpolation library(zoo) theme_set(theme_bw()) ``` ] --- ## Tsibble objects * Like a dataframe or a tibble but geared for time series * index : the time variable - here `year` - but can be day, minute etc... * key : one or several column that corresponding to different time series - here `Country` .left-code[ ```r library(tsibbledata) global_economy ``` ``` # A tsibble: 15,150 x 9 [1Y] # Key: Country [263] Country Code Year GDP Growth CPI Imports Exports Population <fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 Afghanistan AFG 1960 537777811. NA NA 7.02 4.13 8996351 2 Afghanistan AFG 1961 548888896. NA NA 8.10 4.45 9166764 3 Afghanistan AFG 1962 546666678. NA NA 9.35 4.88 9345868 4 Afghanistan AFG 1963 751111191. NA NA 16.9 9.17 9533954 5 Afghanistan AFG 1964 800000044. NA NA 18.1 8.89 9731361 6 Afghanistan AFG 1965 1006666638. NA NA 21.4 11.3 9938414 7 Afghanistan AFG 1966 1399999967. NA NA 18.6 8.57 10152331 8 Afghanistan AFG 1967 1673333418. NA NA 14.2 6.77 10372630 9 Afghanistan AFG 1968 1373333367. NA NA 15.2 8.90 10604346 10 Afghanistan AFG 1969 1408888922. NA NA 15.0 10.1 10854428 # ... with 15,140 more rows ``` ] .right-plot[ ```r key(global_economy) ``` ``` [[1]] Country ``` ] --- ## Tsibble objects * A series with 2 keys .left-code[ ```r olympic_running ``` ``` # A tsibble: 312 x 4 [4Y] # Key: Length, Sex [14] Year Length Sex Time <dbl> <fct> <chr> <dbl> 1 1896 100m men 12 2 1900 100m men 11 3 1904 100m men 11 4 1908 100m men 10.8 5 1912 100m men 10.8 6 1916 100m men NA 7 1920 100m men 10.8 8 1924 100m men 10.6 9 1928 100m men 10.8 10 1932 100m men 10.3 # ... with 302 more rows ``` ] .right-plot[ ```r key(olympic_running) ``` ``` [[1]] Length [[2]] Sex ``` ] --- layout: true # Marine phytoplankton bloom (_Synechoccus_) --- background-image: url(img/2016-Hunter-title.png), url(img/2016-Hunter-time-series.png) background-position: right 20px top 20px, left 100px top 100px background-size: 30%, 80% --- layout: true # Create a tsibble --- ## Read file ```r syn <- read_tsv("data/Syn_temp_light_data.txt", na = c("", "NaN")) ``` ``` Parsed with column specification: cols( Date = col_character(), `Daily Avg. Synechococcus (cells/mL)` = col_double(), `Division rate (1/d)` = col_double(), `Loss rate (1/d)` = col_double(), `Net growth rate (1/d)` = col_double(), `Daily minimum mode of cell volume (um^3)` = col_double(), `PE fluorescence at dawn` = col_double(), `PE fluorescence at dawn normalized by cell volume at dawn (1/um^3)` = col_double(), `Temperature (degree Celsius)` = col_double(), `Daily incident radiation (MJ/um^2)` = col_double() ) ``` --- ## Columns * Many columns have names not compatible with R * No data of interest in first rows and last rows * We are going to concentrate on only 4 columns: * temperature * light * _Synechococcus_ concentration <table class="table table-striped table-hover table-condensed" style="font-size: 9px; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Date </th> <th style="text-align:right;"> Daily Avg. Synechococcus (cells/mL) </th> <th style="text-align:right;"> Division rate (1/d) </th> <th style="text-align:right;"> Loss rate (1/d) </th> <th style="text-align:right;"> Net growth rate (1/d) </th> <th style="text-align:right;"> Daily minimum mode of cell volume (um^3) </th> <th style="text-align:right;"> PE fluorescence at dawn </th> <th style="text-align:right;"> PE fluorescence at dawn normalized by cell volume at dawn (1/um^3) </th> <th style="text-align:right;"> Temperature (degree Celsius) </th> <th style="text-align:right;"> Daily incident radiation (MJ/um^2) </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 01-Jan-2003 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 4.21 </td> <td style="text-align:right;"> 2.11 </td> </tr> <tr> <td style="text-align:left;"> 02-Jan-2003 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 4.06 </td> <td style="text-align:right;"> 1.84 </td> </tr> <tr> <td style="text-align:left;"> 03-Jan-2003 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 3.68 </td> <td style="text-align:right;"> 1.53 </td> </tr> <tr> <td style="text-align:left;"> 04-Jan-2003 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 3.74 </td> <td style="text-align:right;"> 3.38 </td> </tr> <tr> <td style="text-align:left;"> 05-Jan-2003 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 3.82 </td> <td style="text-align:right;"> 8.14 </td> </tr> </tbody> </table> --- ## Rename and select columns then filter ```r syn <- syn %>% mutate(Date = lubridate::dmy(Date)) %>% rename(date = Date, temperature=`Temperature (degree Celsius)`, syn_ml = `Daily Avg. Synechococcus (cells/mL)`, radiation= `Daily incident radiation (MJ/um^2)`) %>% select(date, syn_ml, temperature, radiation) %>% filter(date > lubridate::ymd("2003-05-28") & date < lubridate::ymd("2018-09-04")) ``` <table class="table table-striped table-hover table-condensed" style="font-size: 9px; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> date </th> <th style="text-align:right;"> syn_ml </th> <th style="text-align:right;"> temperature </th> <th style="text-align:right;"> radiation </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> 2003-05-29 </td> <td style="text-align:right;"> 3344.51 </td> <td style="text-align:right;"> 11.95 </td> <td style="text-align:right;"> 22.83 </td> </tr> <tr> <td style="text-align:left;"> 2003-05-30 </td> <td style="text-align:right;"> 3521.90 </td> <td style="text-align:right;"> 12.00 </td> <td style="text-align:right;"> 28.28 </td> </tr> <tr> <td style="text-align:left;"> 2003-05-31 </td> <td style="text-align:right;"> 3939.43 </td> <td style="text-align:right;"> 11.92 </td> <td style="text-align:right;"> 19.40 </td> </tr> <tr> <td style="text-align:left;"> 2003-06-01 </td> <td style="text-align:right;"> 4796.46 </td> <td style="text-align:right;"> 11.70 </td> <td style="text-align:right;"> </td> </tr> <tr> <td style="text-align:left;"> 2003-06-02 </td> <td style="text-align:right;"> 4348.95 </td> <td style="text-align:right;"> 11.55 </td> <td style="text-align:right;"> </td> </tr> </tbody> </table> --- ## Transform to long form and create tsibble .pull-left[ ```r syn_long <- syn %>% pivot_longer(-date, names_to = "parameter", values_to = "value" ) tsyn <- tsibble::as_tsibble(syn_long, index = date, key = parameter, regular = TRUE, validate = TRUE) ``` ] .pull-right[ ``` # A tsibble: 16,731 x 3 [1D] # Key: parameter [3] date parameter value <date> <chr> <dbl> 1 2003-05-29 radiation 22.8 2 2003-05-30 radiation 28.3 3 2003-05-31 radiation 19.4 4 2003-06-01 radiation NA 5 2003-06-02 radiation NA 6 2003-06-03 radiation NA 7 2003-06-04 radiation NA 8 2003-06-05 radiation 10.6 9 2003-06-06 radiation 29.6 10 2003-06-07 radiation NA # ... with 16,721 more rows ``` ] --- layout: true # Plot data --- ## Use autoplot .pull-left[ ```r autoplot(tsyn) + xlab("Date") + ylab("Value") + scale_x_date(date_breaks="1 year", date_labels = "%Y") + facet_grid(rows = vars(parameter), scales= "free_y") ``` * Can also use ggplot2 of course ] .pull-right[ 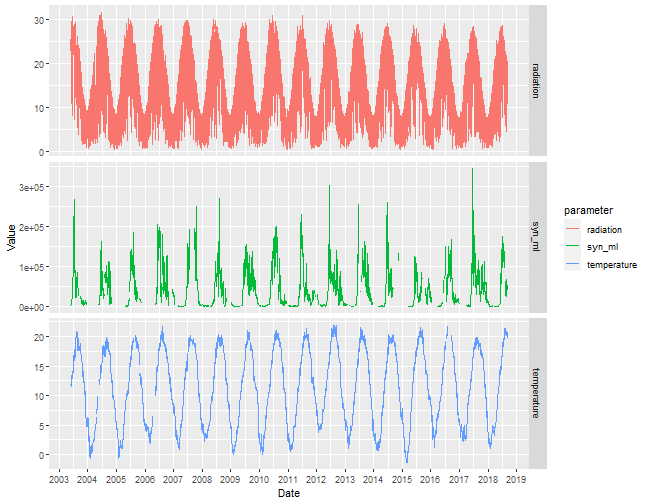<!-- --> ] --- ## Tiles ```r tsyn %>% filter( parameter == "temperature") %>% ggplot(aes(x = date, y = 1)) + geom_tile(aes(fill = value)) + scale_fill_gradient2(low = "navy", mid = "yellow", high = "red", midpoint = 28) + ylab("") + scale_y_discrete(expand = c(0, 0)) + scale_x_date(date_breaks="1 year", date_labels = "%Y") ``` 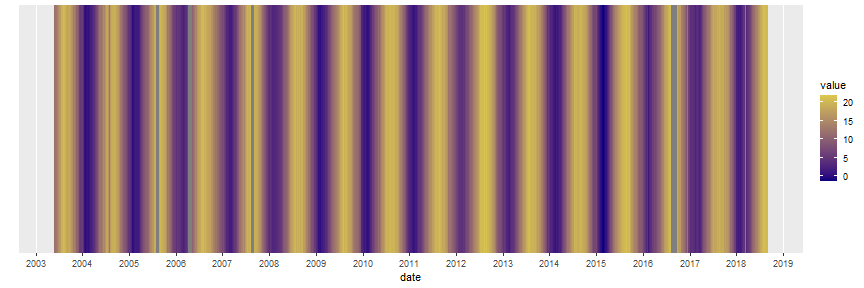<!-- --> --- ## Season plots For all variables .pull-left[ ```r tsyn %>% feasts::gg_season(y=value, period="year") ``` ] .pull-right[ <!-- --> ] --- ## Season plots For cell concentration better to use log scale .pull-left[ ```r tsyn %>% filter(parameter == "syn_ml") %>% feasts::gg_season(y=value, period="year") + scale_y_log10() ``` ] .pull-right[ <!-- --> ] --- ## Focus on one year .pull-left[ ```r tsyn %>% filter(year(date) == 2010) %>% autoplot() + xlab("Date") + ylab("Value") + scale_x_date(date_breaks="1 month", date_labels = "%b") + facet_grid(rows = vars(parameter), scales= "free_y") + ggtitle("Year 2010") ``` .warning[ Note holes in data. These holes must be filled before time series analysis] ] .pull-right[ <!-- --> ] --- layout: true # Cleaning and smoothing the data --- ## Estimating missing values .pull-left[ Package `zoo` * zoo::na.approx() - Linear interpolation * zoo::na.spline() - Spline interpolation ```r tsyn_filled <- tsyn %>% # filter(parameter == "temperature") %>% tsibble::group_by_key()%>% mutate(value = zoo::na.approx(value, na.rm = FALSE)) ``` ] .pull-right[ <!-- --> ] --- ## Moving average .pull-left[ `\(\hat{y}_{t} = \frac{1}{2k+1} \sum_{j=-k}^k y_{t+j}\)` * All points have same weight * Smoothing increases with `\(k\)` * Package `slider` is currently developed ```r tsyn_MA <- tsyn_filled %>% group_by_key() %>% mutate(value_MA = tsibble::slide_dbl(value, mean, na.rm = TRUE, .size = 25, .align = "center")) ``` ] .pull-right[ <!-- --> ] --- ## Exponential smoothing .pull-left[ `\(\hat{y}_{T+1|T} = \alpha y_T + \alpha(1-\alpha) y_{T-1} + \alpha(1-\alpha)^2 y_{T-2}+ \cdots\)` `\(0 \le \alpha \le 1\)` * Forecasting method (use points from past only) * Recent points have more weight * Smoothing increases with `\(\alpha\)` decreasing ```r tsyn_ETS <- tsyn_filled %>% model(fable::ETS(value ~ season (period = "1 year") + trend(alpha = 0.3))) ``` <table class="table table-striped table-hover table-condensed" style="font-size: 9px; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> parameter </th> <th style="text-align:left;"> .model </th> <th style="text-align:left;"> date </th> <th style="text-align:right;"> value </th> <th style="text-align:right;"> level </th> <th style="text-align:right;"> slope </th> <th style="text-align:right;"> remainder </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> radiation </td> <td style="text-align:left;"> fable::ETS(value ~ season(period = "1 year") + trend(alpha = 0.3)) </td> <td style="text-align:left;"> 2003-05-28 </td> <td style="text-align:right;"> </td> <td style="text-align:right;"> 19.89920 </td> <td style="text-align:right;"> 0.5388970 </td> <td style="text-align:right;"> </td> </tr> <tr> <td style="text-align:left;"> radiation </td> <td style="text-align:left;"> fable::ETS(value ~ season(period = "1 year") + trend(alpha = 0.3)) </td> <td style="text-align:left;"> 2003-05-29 </td> <td style="text-align:right;"> 22.83 </td> <td style="text-align:right;"> 21.15567 </td> <td style="text-align:right;"> 0.5391361 </td> <td style="text-align:right;"> 0.1170316 </td> </tr> <tr> <td style="text-align:left;"> radiation </td> <td style="text-align:left;"> fable::ETS(value ~ season(period = "1 year") + trend(alpha = 0.3)) </td> <td style="text-align:left;"> 2003-05-30 </td> <td style="text-align:right;"> 28.28 </td> <td style="text-align:right;"> 23.67036 </td> <td style="text-align:right;"> 0.5397947 </td> <td style="text-align:right;"> 0.3035379 </td> </tr> </tbody> </table> ] .pull-right[ <!-- --> ] --- layout: true # Determining periodicity --- ## Autocorrelation .pull-left[ `\(r_{k} = \frac{\sum\limits_{t=k+1}^T (y_{t}-\bar{y})(y_{t-k}-\bar{y})}{\sum\limits_{t=1}^T (y_{t}-\bar{y})^2}\)` Use function `feasts::gg_lag` to visualize * For temperature ```r tsyn %>% filter(parameter == "temperature") %>% feasts::gg_lag(lags=c(90, 182, 365), geom = "point") ``` ] .pull-right[ <!-- --> ] --- ## Autocorrelation .pull-left[ Use function `feasts::ACF` to compute ```r tsyn_filled %>% feasts::ACF(value, lag_max = 500) %>% autoplot() + scale_x_continuous(breaks=seq(0,500, by=100)) ``` ] .pull-right[ <!-- --> ] --- ## Cross correlation .pull-left[ * Same as autocorrelation but now between 2 series (light and temperature) * Use function `feasts::CCF` * Go from long form to wide form because variables must be in 2 different columns ```r lag_max = 400 tsyn_filled %>% pivot_wider(names_from = "parameter", values_from = "value") %>% as_tsibble(index = date) %>% feasts::CCF(temperature, radiation, lag_max = lag_max) %>% tibble::rowid_to_column(var = "lag2") %>% mutate(lag2 = lag2 - lag_max) %>% ggplot() + geom_point(aes(x=lag2, y = ccf)) + scale_x_continuous(breaks=seq(-lag_max,lag_max, by=50)) + ggtitle("Temperature vs radiation") ``` ] .pull-right[ <!-- --> ] --- ## STL decomposition .pull-left[ Three elements: season + trend + error * Use function `feasts::STL` * For temperature ```r dcmp_temp <-tsyn_filled %>% filter(parameter == "temperature") %>% model(stl = feasts::STL(value ~ season(period= "1 year", window = 11), robust = TRUE)) ``` <table class="table table-striped table-hover table-condensed" style="font-size: 9px; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> parameter </th> <th style="text-align:left;"> .model </th> <th style="text-align:left;"> date </th> <th style="text-align:right;"> value </th> <th style="text-align:right;"> trend </th> <th style="text-align:right;"> season_1 year </th> <th style="text-align:right;"> remainder </th> <th style="text-align:right;"> season_adjust </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> temperature </td> <td style="text-align:left;"> stl </td> <td style="text-align:left;"> 2003-05-29 </td> <td style="text-align:right;"> 11.95 </td> <td style="text-align:right;"> 10.30981 </td> <td style="text-align:right;"> 1.482655 </td> <td style="text-align:right;"> 0.1575318 </td> <td style="text-align:right;"> 10.46735 </td> </tr> <tr> <td style="text-align:left;"> temperature </td> <td style="text-align:left;"> stl </td> <td style="text-align:left;"> 2003-05-30 </td> <td style="text-align:right;"> 12.00 </td> <td style="text-align:right;"> 10.31027 </td> <td style="text-align:right;"> 1.726744 </td> <td style="text-align:right;"> -0.0370158 </td> <td style="text-align:right;"> 10.27326 </td> </tr> <tr> <td style="text-align:left;"> temperature </td> <td style="text-align:left;"> stl </td> <td style="text-align:left;"> 2003-05-31 </td> <td style="text-align:right;"> 11.92 </td> <td style="text-align:right;"> 10.31073 </td> <td style="text-align:right;"> 1.812566 </td> <td style="text-align:right;"> -0.2032953 </td> <td style="text-align:right;"> 10.10743 </td> </tr> </tbody> </table> ] .pull-right[ <!-- --> ] --- ## Wavelet analysis .pull-left[ Extension of Fourier analysis https://en.wikipedia.org/wiki/Wavelet Package `WaveletComp` http://www.hs-stat.com/projects/WaveletComp/WaveletComp_guided_tour.pdf ] .pull-right[ <!-- --> ] --- ## Wavelet analysis .pull-left[ ### For temperature ```r tsyn_wavelet <- tsyn_filled %>% filter(parameter == "temperature") %>% WaveletComp::analyze.wavelet( my.series = "value", loess.span = 0, dt = 1/365, dj = 1/20, lowerPeriod = 1/18, upperPeriod = 4, make.pval = TRUE, n.sim = 10) ``` ] .pull-right[ <!-- --> ] --- ## Wavelet analysis .pull-left[ ### For _Synechococcus_ ```r tsyn_wavelet <- tsyn_filled %>% filter(parameter == "syn_ml") %>% mutate(value = log10(value)) %>% tidyr::fill(value, .direction = "down") %>% WaveletComp::analyze.wavelet( my.series = "value", loess.span = 0, dt = 1/365, dj = 1/20, lowerPeriod = 1/18, upperPeriod = 4, make.pval = TRUE, n.sim = 10) ``` ] .pull-right[ <!-- --> ] --- layout: false class: middle, inverse # Recap ## What we saw today .font150[ * Time series in environmental sciences * Manipulate dates * Create tsibble objects * Visualize time series * Smooth time series * Determine periodicity and trends ] ## Topics to explore on your own .font150[ * Modelling * Forecasting * Wavelet analysis ]